Deep Learning for Analysis of Imbalanced Medical Image Datasets
- 0 Collaborators
Medical image datasets are predominantly composed of “normal” samples with only a small percentage of “abnormal” ones, leading to the so-called class imbalance problem. It has been established that class imbalance can have significant detrimental effect on training of machine learning classifiers. In such cases, classifiers typically focus on learning the large classes – thereby leading to poor classification accuracy for the smaller classes. However, in case of medical diagnosis, misclassification costs are often unequal and classifying the minority (image containing cancerous tumor) samples as majority (images for healthy patient or the tumor is benign) implies serious consequences. Additionally, the distribution of test data may differ from that of the learning samples and the true misclassification costs may be unknown at learning time. Although much awareness of the issues related to data imbalance has been raised, many of the key problems still remain open and are in fact encountered more often, especially when applied to medical image datasets. In classical machine learning, class imbalance is well addressed and tackled at the data level or at the level of the classifier. Although deep learning has recently achieved great success due to its high learning capacity and its ability to automatically learn accurate underlying descriptors of input data, but it still cannot escape from such negative impact of imbalanced data. The techniques used to handle class imbalance in case of classical machine learning or ‘shallow’ models, are not applicable for deep Convolutional Neural Networks (ConvNets) applications on medical image datasets. While the issue is well addressed in traditional machine learning algorithms, no research on this issue for deep networks (with application to real medical imaging datasets) is available in the literature. In this project, supported by the Intel AI research grant, we will study the impact of class imbalance on the performance of ConvNets for the three main medical image analysis problems viz., (i) disease or abnormality detection, (ii) region of interest segmentation (iii) disease classification from real medical image datasets. It is worth mentioning that most of the study reported in the literature in this field used synthetic datasets or dataset acquired in a controlled environment. Our plan, on the other hand, is to investigate the effect of class imbalance in case of publicly available large heterogeneous biomedical image datasets of real patient, in the presence of other commonly occurring limitations in clinical data such as, limited training sample size, large number of features, correlation among extracted features, and noise. The developed models will be implemented using Intel® AI DevCloud (Intel Optimized version of Tensorflow and keras in Intel distribution for Python). ...learn more
Project status: Under Development
Intel Technologies
AI DevCloud / Xeon,
MKL,
Intel Opt ML/DL Framework,
BigDL,
Movidius NCS
Overview / Usage
Class imbalance can take many forms, particularly in the context of multiclass classification, for ConvNets. In some problems
only one class might be under-represented or over-represented, while in other case every class may have a different number of
examples. In this project we will first study the impact of class imbalance on the performance of ConvNets for the three main
medical image analysis problems viz., (i) disease or abnormality detection, (ii) region of interest segmentation (iii) disease
classification from real medical image datasets. This is worth mentioning that most of the study reported in the literature in this
field used synthetic datasets or dataset acquired in a controlled environment. Whereas our plan is to investigate the effect of class
imbalance in case of publicly available large heterogeneous biomedical image datasets of real patient, in the presence of the other
commonly occurring limitations in clinical data such as, limited training sample size, large number of features, correlation among
extracted features, and noise.
In this project, we plan to used three large datasets (suitable for deep learning) of three different modalities (X-ray, MRI, and Color
Fundus Photography) dedicated for each of the three medical image analysis tasks (automated detection of 14 common thorax
disease, segmentation of high and low-grade brain tumors from brain MR images, and classification of diabetic retinopathy into
five classes). The dataset description is given below.
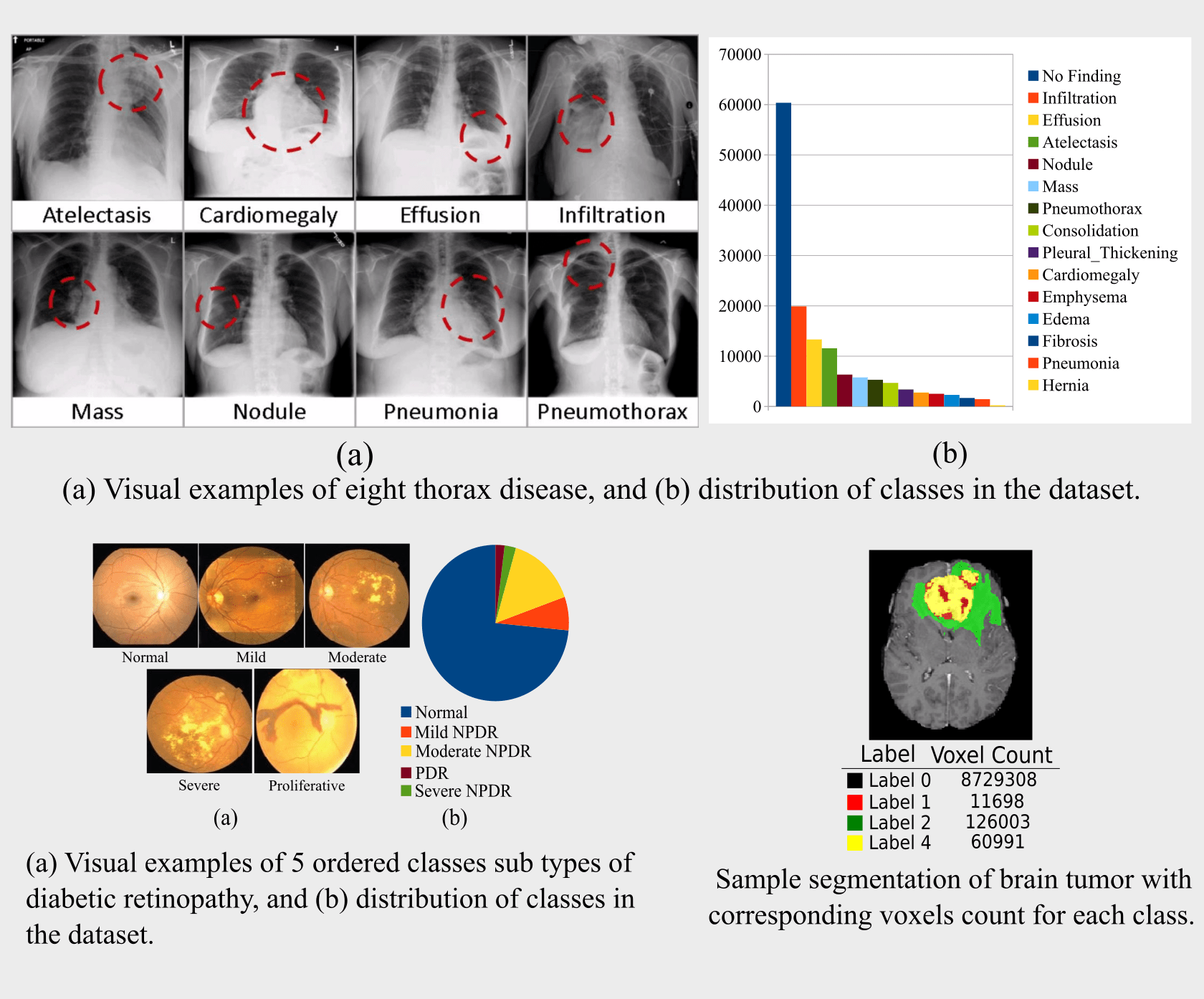
(a) NIH Chest X-ray Dataset of 14 Common Thorax Disease: Chest X-ray is one of the most frequent and cost-effective
medical imaging examination. However clinical diagnosis of chest X-ray can be challenging. This comprises
112,120 frontal-view X-ray images of 30,805 unique patients with fourteen common thoracic pathologies including
Atelectasis, Consolidation, Infiltration, Pneumothorax, Edema, Emphysema, Fibrosis, Effusion, Pneumonia,
Pleural_thickening, Cardiomegaly, Nodule, Mass and Hernia. For each disease image there is one or more labeled
bounding box marked by radiologists. If one image contains multiple disease instances, then each such is localized
separately by labeled bounding boxes. As shown in Fig. 1 the class distribution in the dataset is massively imbalanced,
and this is termed linear imbalance.
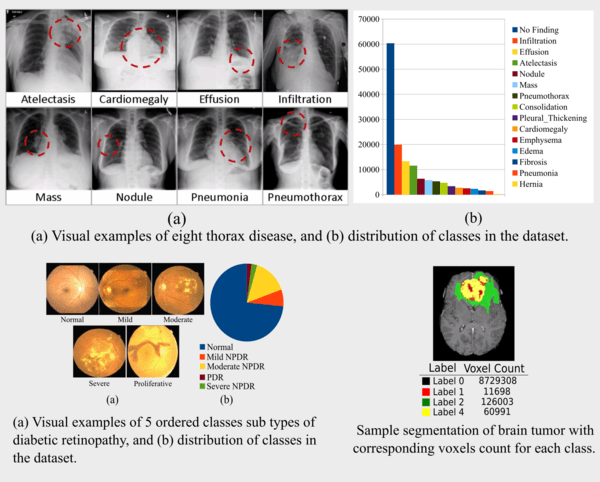
(b) Kaggle Diabetic Retinopathy Dataset: This dataset contains 35126 high-resolution eye images in the training
set divided into 5 fairly unbalanced classes as given in Fig. 2, and the objective is to predict the class (one of the 5
numbers) for each of the 53576 test images in the dataset. The classes imbalance in the dataset can be considered as step
imbalance, where the number of examples is approximately equal within minority classes (Severe NPDR & PDR and
Moderate NPDR & Mild NPDR) and equal within majority class (Normal) but differs between the majority and minority
classes.
(c) MICCAI Brain Tumor Segmentation Challenge Dataset: The dataset consists of 210 HGG and 75 LGG glioma cases. Each patient MRI scan set has four MRI sequences, encompassing native (T1) and post-contrast enhanced T1-weighted (T1C), T2-weighted (T2),
and T2 Fluid-Attenuated Inversion Recovery (FLAIR) volumes having 155 2D slices of 240X240 resolution. Each voxel of the ground truth image(s) is labeled using three labels for the intra tumoral components such as the necrotic and non-enhancing tumor (NCR/NET – label 1), the peritumoral edema (ED – label 2), and the GD-enhancing tumor (ET – label 4), and label 0 for everything else encompassing healthy brain tissue and background (label 3 is not used). The task is to segment each patient MRI scan into three sub-regions: (i) the “enhancing tumor” (ET) (label 4), (ii) the “tumor core” (TC) (label 1 + label 4), and (iii) the “whole tumor” (WT) (label 1 + label 2 + label 4). The best performing method achieved DICE score of 72.9, 78.5, 88.6 for the ET, TC, and WT. Although it is pretty easy to learn to segment the enhancing tumor due to the contrast enhancement, the best performing segmentation model could not attain high accuracy for this task. Since voxels belonging to the ET are very few as compared to
those belonging to the whole tumor class, the classifier learning of features from the minority class is poor.
Methodology / Approach
will design three strategies for handling ConvNet training on medical image datasets having imbalanced classes. These are elaborated below.
(a) Cost sensitive learning: We will first experiment with the three standard loss functions i.e., Hinge Loss, Euclidean Loss and traditional Cross Entropy Loss for the regression task (localization of thoracic diseases) and the traditional softmax loss for the multi-class classification task (Diabetic Retinopathy classification and patch-based brain tumor segmentation). Since all these three datasets have an unbalanced numbers of pathology and normal classes, the models will have difficulty in learning positive instances (images with pathologies) and use of the traditional loss functions that treat misclassification of each class equally will lead to poor performance. So, we would like to propose and use a weighted version of these loss functions by introducing positive/negative balancing factor to enforce the learning of positive examples. And comparisons will be made between the performance of the networks with the weighted and traditional loss functions. This could be thought of as Cost sensitive learning that assigns different cost to misclassification of examples from different classes. Another adaptation of neural network to be cost sensitive is to modify the learning rate, such that higher cost examples contribute more to the update of weights. And finally we can train the network by minimizing the misclassification cost instead of the standard loss function.
(b) One-class learning: This strategy involves training a single classifier per class, with the samples of that class considered as positive and all other samples as negatives. This is a concept learning technique that learns to recognize positive instances rather than discriminating between two classes. We will use deep Autoencoders for this purpose and will train them to perform autoassociative mapping, i.e. identity function. Then, the classification of a new example is to be made based on a reconstruction error between the input and output patterns, e.g. absolute error, squared sum of errors, Euclidean or Mahalanobis distance. This will be a novel application of Autoencoders for deep learning on imbalanced datasets.
(c) Two-phase training with pre-training on randomly oversampled/undersampled dataset: We will examine two variants of two-phase training method, one on oversampled and the other on undersampled dataset. First we will try to balance the datasets using commonly used methods of over sampling through data augmentation or undersampling through elimination of redundant examples close to the boundary between classes. This is to be followed by pre-training of the ConvNet on balanced dataset, and finally fine-tuning the last output layer before softmax on the original, imbalanced dataset (by keeping the same hyperparameters and learning rate decay policy as in the first phase). We will test the effects of class imbalance on classification performance and compare with the three proposed methods to address the issue. The metric that is most widely used to evaluate a classifier performance in the context of multiclass classification with ConvNets is overall accuracy, which is the proportion of test examples that were correctly classified. However, it has some significant and long acknowledged limitations, particularly in the context of imbalanced datasets. Specifically, when the test set is imbalanced, accuracy will favor classes that are over-represented in some cases leading to highly misleading assessment. Another issue that may arise when the test set is balanced but the training set is imbalanced. This might result in a situation when a decision threshold is moved to reflect the estimated class prior probabilities and cause a low accuracy measure in the test set, while the true discriminative power of the classifier does not change.
Technologies Used
Intel® Xeon® Scalable Processor
Intel® Movidius™ Neural Compute Stick
Intel® AI DevCloud
Intel® Distribution for Python*
Intel® Optimization for TensorFlow*
Intel® Optimization for Keras*
Intel® Optimization for Theano*
Library for Deep Neural Networks (Intel® MKL-DNN)
BigDL
Insight Segmentation and Registration Toolkit (ITK)
OpenCV